We are proud to share our interdisciplinary and collaborative work at the Department of Electrical and Electronics Engineering and UMRAM of Bilkent University and the School of Medicine of Koç University on single-cell RNA sequencing (scRNA-seq) with graph- and transformer-based neural networks!
Our article is now published in the Special Issue on Learning on Graphs for Biology and Medicine of the IEEE Transactions on Signal and Information Processing over Networks (IEEE TSIPN)!
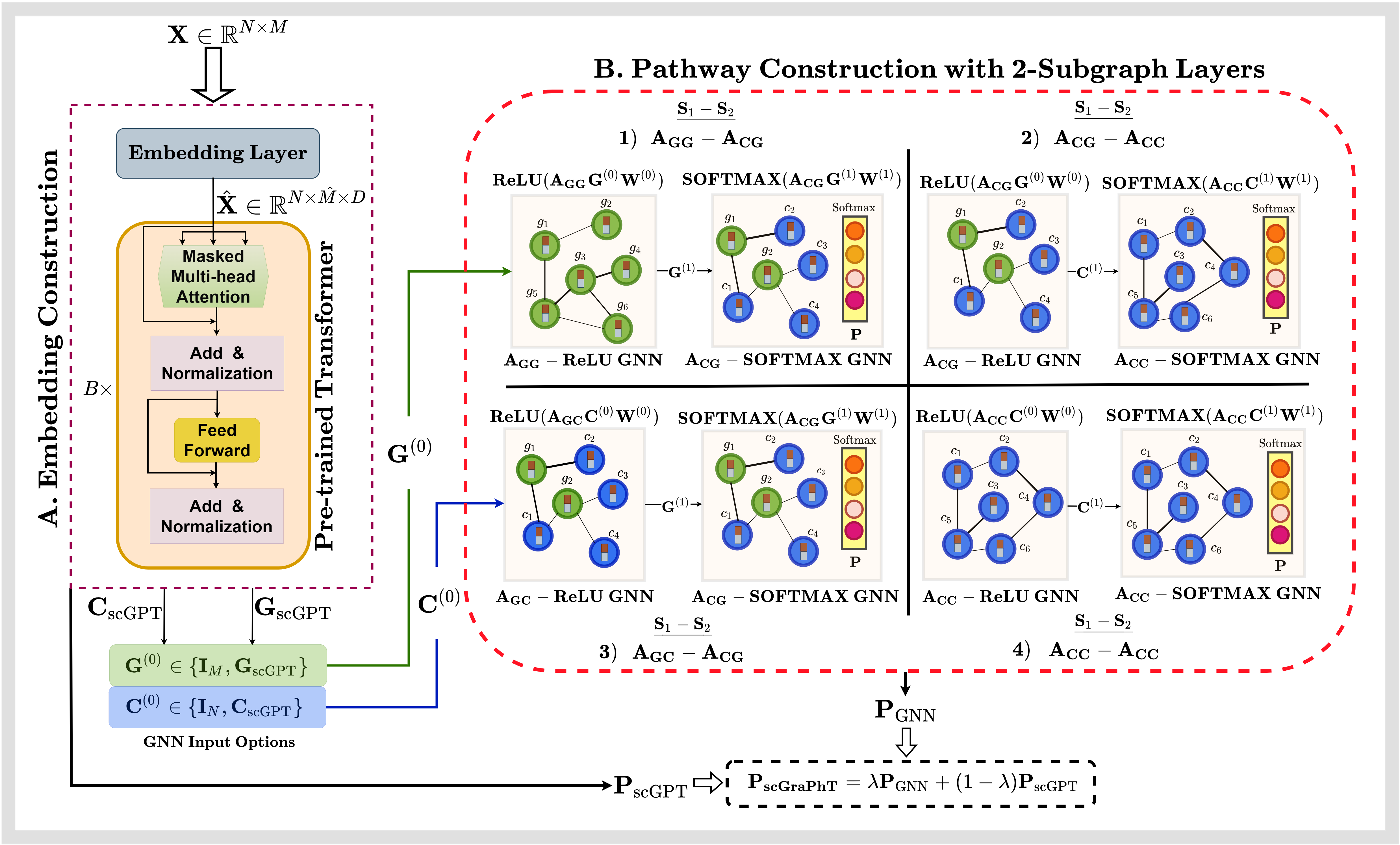
In this article, we introduce scGraPhT, a graph- and transformer-based cell type annotation method for scRNA-seq, which is a state-of-the-art technique to examine gene expression profiles at the individual cell level, paving the way for significant discoveries in research fields such as pathology, immunology, cancer, genomics, and regenerative medicine.
Our approach, scGraPhT, allows integrating pre-trained transformers to extract rich representations of scRNA-seq data with a multi-layered graph neural network (GNN) to capture cell-cell and cell-gene relationships. scGrapPhT brings together the power of transformers that excel in modeling local contextual information and GNNs excelling in modeling global structural relationships.
Moreover, scGraPhT also:
– can work with both homogeneous and heterogeneous relationships through subgraph layers to offer comprehensive assessment,
– does not require exhaustive pre-training stages by being able to leverage the readily available existing pre-trained works as the initial point,
– outperforms existing works regarding model performance on the standard scRNA-seq benchmark datasets.
For more details and code:
Paper: https://ieeexplore.ieee.org/document/11015257
Code: https://github.com/koc-lab/scgrapht
Abstract:
The invention of single-cell RNA sequencing (scRNA-seq) has enabled transcriptomic examination of cells on an individual basis, uncovering cell-to-cell phenotypic heterogeneity within isogenic cell populations and tissues. Inevitably, cell type annotation has emerged as a fundamental, albeit challenging task in scRNA-seq data analysis, which involves identifying and characterizing cells based on their unique molecular profiles. Recently, deep learning techniques with their data-driven priors have shown significant promise in this task. On the one hand, task-agnostic transformer networks pre-trained on large-scale biological databases can capture generalizable data representations to serve as foundation models despite their ineffectiveness in characterizing intricate relationships between biological entities such as cells or genes. Contrarily, task-specific graph neural networks (GNNs) can be trained on target datasets to sensitively characterize entity relationships, but they can suffer from relatively poor generalizability. Furthermore, existing GNNs focus exclusively on either homogeneous or heterogeneous relationships, limiting their ability to offer a complete picture of the diverse inner structure of cells. In this study, we propose a novel merged transformer-graph model, scGraPhT, that integrates a pre-trained transformer to extract rich representations of scRNA-seq data with a multi-layered GNN to capture cell-cell, cell-gene, and gene-gene relationships. Different from previous GNNs, scGraPhT examines both homogeneous and heterogeneous relationships through subgraph layers to offer a more comprehensive assessment. Since the graph construction in scGraPhT relies on representations from a pre-trained transformer model, our approach does not require costly training procedures. Moreover, scGraPhT can also be adapted to leverage any transformer-based single-cell annotation method, such as scGPT or scBERT, that produces suitable embedding representations as input for GNNs. Demonstrations on three scRNA-seq benchmark datasets indicate that scGraPhT outperforms state-of-the-art annotation methods without compromising efficiency. We offer insights into performance improvements by employing Grad-CAM, a visual explainability method that elucidates the complementary nature of the GNN- and transformer-based components of scGraPhT to improve its predictive performance. We share our publicly available source codes and datasets for reproducibility.